Systolic Accelerator for Molecular Biological Applications
History
The design of SAMBA was decided in september 1993. The main decision was to choose the target technology: programmable or custom architecture. It was decided to design a systolic array with fully dedicated processors to get high performance and minimize the design of a programming environment – a task which usually requires a lot of efforts.
Algorithm
The algorithm implemented on SAMBA belongs to the dynamic programming class. The recurrence equation comes from the Smith and Waterman algorithm. However, it has been parametrized to cover a larger scope. A similarity matrix is calculated recursively using the following equation:
-
H(i,j)=max(delta,E(i,j),F(i,j),H)i-1,j-1)+sub(S1i,S2j))
with:
-
E(i,j) = Max (H(i,j-1)-alpha,E(i,j-1)-beta)
F(i,j) = Max (H(i-1,j)-alpha,F(i-1,j)-beta)
and the initializations given by:
-
H(i,0) = E(i,0) = hi(i)
H(0,j) = F(0,j) = vi(j)
alpha, beta, delta, hi and vi are parameters for tuning the algorithm to local or global search, with or without gap penalty
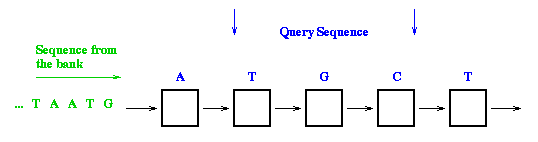
Parallelization
The sequence comparison on a linear systolic array proceeds as follows: one sequence (the query sequence) is stored into the array (one character per processor) and the other sequence flows from the left to right through the array. During each systolic step, one elementary matrix computation is performed on each processor. The result is available on the rightmost processor when the last character of the flowing sequence is output.
(N x M) / N + M – 1 = N (considering that M >> N)
N = size of the query sequence (number of processors)
M = size of the database
Architecture
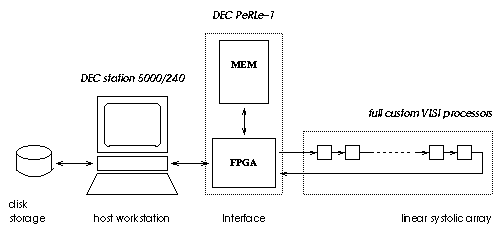
SAMBA comprised a workstation (with its local disk), a systolic array made out of 128 VLSI full custom processors and a FPGA-Memory interface which fills the gap between a complete hardwired array of processors and a programmable Von Neuman machine.

Bibliography
- D. Lavenier, Speeding up genome computations with a systolic accelerator, SIAM news, 31 (8) 1998 (pdf)
- P. Guerdoux-Jamet, D. Lavenier, SAMBA: Hardware Accelerator for Biological Sequence Comparison,
CABIOS, 13 (6) 1997. - D. Lavenier, Dedicated Hardware for Biological Sequence Comparison, Journal of Universal Computer Science, 2 (2) 1996 (pdf)
- P. Guerdoux-Jamet, D. Lavenier, C. Wagner, P. Quinton, Design and Implementation of a Parallel Architecture for Biological Sequence Comparison, EuroPar’96, European Conference on Parallelism, Lyon, France, 1996. (pdf)
